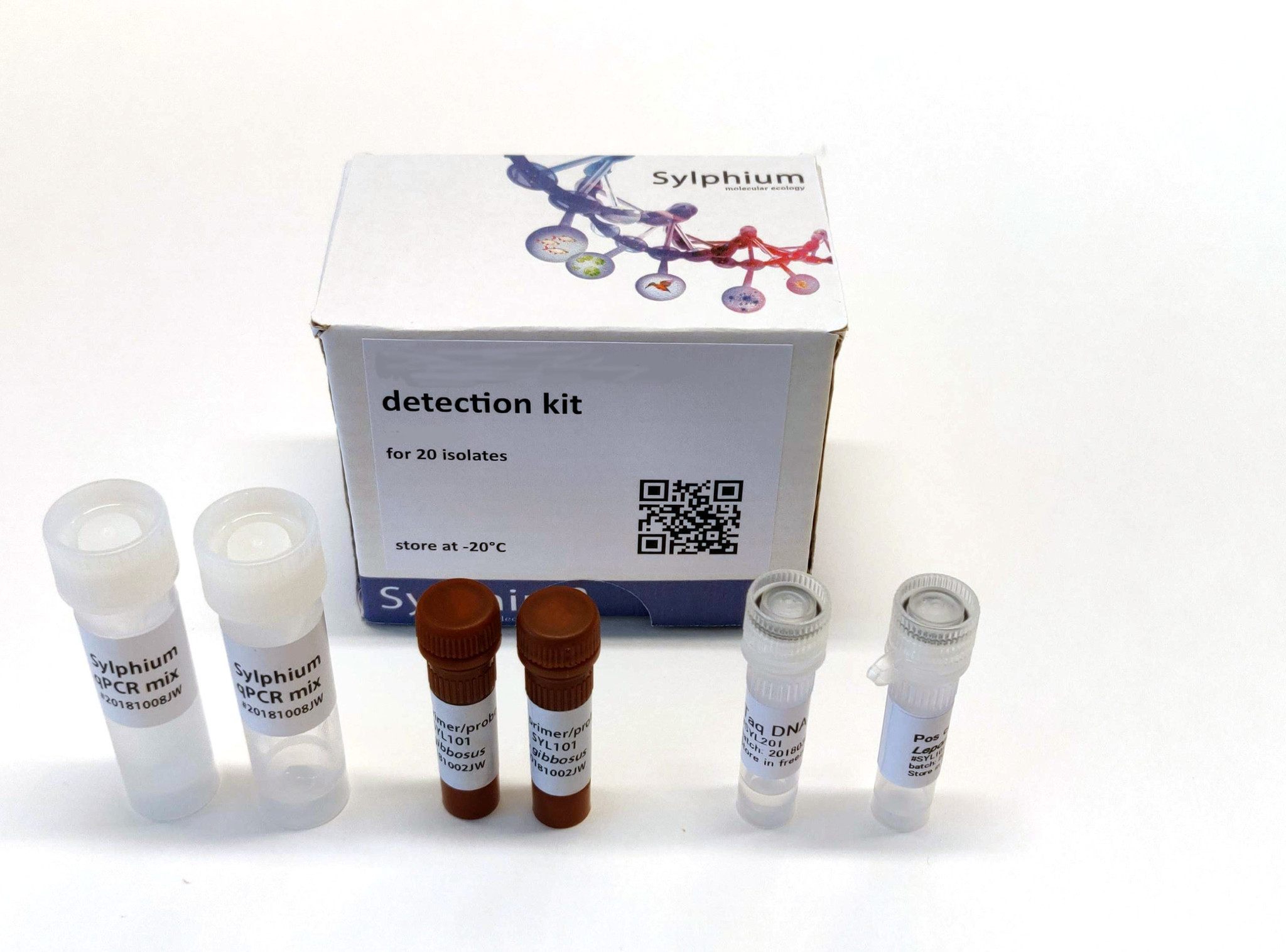
The Triturus cristatus (great crested newt) eDNA detection kit is able to detect the amfibian species Triturus cristatus in all water samples worldwide and contains enough materials for 200 reactions. The kit is based on the fast, sensitive, and proven primers/probe qPCR technique. The kit contains primers and a probe for detecting a highly specific sequence present on the mitochondrial DNA of the species Triturus cristatus.
Species information
The northern crested newt, also known as the great crested newt or warty newt (Triturus cristatus) is a newt in the family Salamandridae. The range of the northern crested newt covers most of Europe, although the species is not found in Southern Europe. The northern edge of its range runs through northern France, Great Britain and southern Scandinavia to the Republic of Karelia in Russia; the southern margin runs through central France, southwest Romania, Moldavia and Ukraine, heading from there into central Russia and through the Ural Mountains. The eastern extent of the great crested newt’s range reaches into Western Siberia, running from the Perm Krai to the Kurgan Oblast.
Outside of the breeding season, great crested newts are forest-dwellers and inhabit coniferous, mixed and deciduous forests, as well as bushland, meadows, and parks and gardens. In the southern extent of their range and on the edges of steppe environments, they also inhabit insular forests.
In English: Northern crested newt or great crested newt
In German: Nördlicher Kammmolch
In French: Triton crêté
In Dutch: Kamsalamander
Primer design
The primers and probe are specially designed to be used with eDNA samples and have the following properties:
- Highest possible sensitivity (1 DNA copy per reaction). Environmental water samples contain normally very low amounts of target DNA.
- Strong fluorescence signal with low background noise. Isolated environmental samples contain residues of naturally occurring auto fluoresce substances that will interfere with the measurements. A strong fluorescence signal from the analyses is required for these kind of samples.
- 100% specificity. Isolated DNA from environmental samples contains billions of DNA fragments from bacteria, protozoa, plants, animals, etc. Not only animals from the same order must be taken in account during primer/probe design, but all known DNA sequences must be checked for nonspecific binding of the primers and probe. This is validated by experimental and bioinformatical studies.
The kit is developed and optimized to be used on eDNA isolates purified using the eDNA isolation kit (#SYL002) from Sylphium molecular ecology.
Kit contents
The kit contains materials for 200 reactions.
- Positive control (Triturus cristatus DNA)
- 2x Sylphium qPCR mix (100 reactions) without primers and probes
- 2x Primer/probe mix (100 reactions) for detection of Triturus cristatus (FAM dye)
- 1x Taq DNA polymerase (200 reactions)
- Protocol and primer/probe validation report
Manual